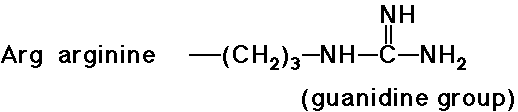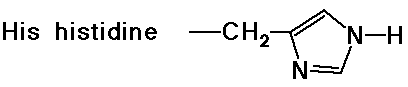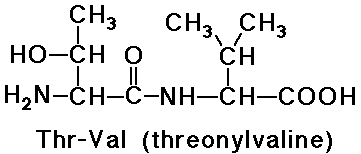Amino Acids & Proteins

Amino Acids
- natural amino acids are alpha (position of the amine) and L (stereochemistry)
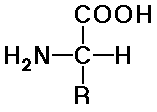
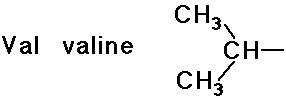
- structures differ by the R groups
there are 20 common natural amino acids
10 amino acids are essential to human nutrition
(we can biosynthesize the others)
Side Chains
- include a great variety of functional groups and properties
- glycine has no side chain
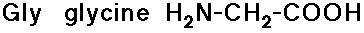
- simple alkyl groups (nonpolar)
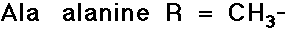
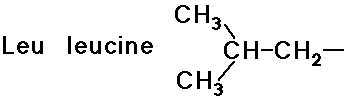
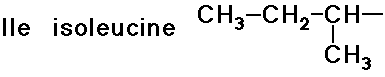
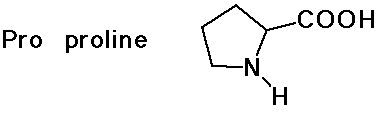
- cyclic side chain (a secondary amine)

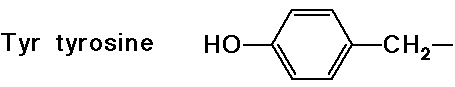
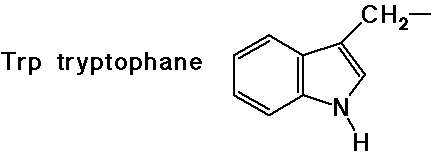
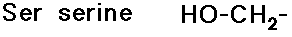
 (also
note that tyrosine is a phenol)
(also
note that tyrosine is a phenol)
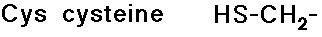
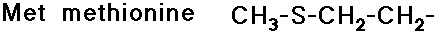
- carboxylic acid side chains

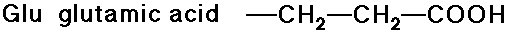
- amide side chains (from the above acids)

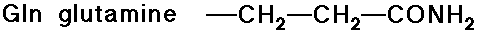
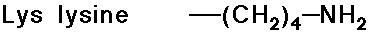
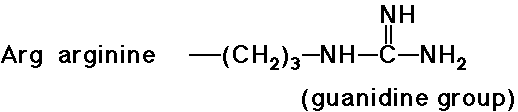
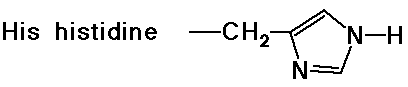
Acid-Base Properties of the Amino Acids
- basic amine group and acidic carboxyl group are typically both in
their ionized forms in aqueous solution around biological pH values
- pKa for the alpha-amino group is about 9-10
- pKa for the carboxylic acid group is about 2-3
- below pH ~ 2, mainly cationic form (ammonium ion)
- between pH ~ 2 - 10, mainly zwitterionic form (both ammonium cation
and carboxylate anion)
- above pH ~ 10, mainly anionic form (carboxylate anion)
- amino acids with side chains that can also ionize have more complicated
behavior
The Isoelectric Point
- pH at which the major form of the molecule is neutral
(and there are equal - but small - amounts of cationic and anionic forms)
- for most amino acids, the isoelectric point is midway between their
two pKa values (the middle of their neutral range)
- for amino acids with ionizable side chains, isoelectric points are
at higher pH (for basic side chains) and lower pH (for acidic side
chains)
Electrophoresis
- amino acids can be separated based on their charge in solution at
a given pH
e.g., at pH 7, alanine is aprox. neutral, arginine is mainly +, glutamic
acid is mainly -
- depending on their charge, the molecules migrate towards either
a positive or negative electrode (moving through wet gel or paper)
Peptides - Amino Acids joined by Amide Bonds
- naming goes from N-terminus (free amine) to C-terminus (free carboxyl)
- -ine ending of the amino acids are replaced by -yl (except the last
one)
e.g., threonylvaline is a dipeptide
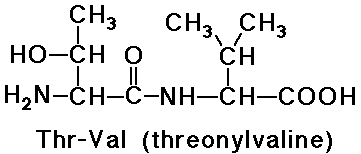
- note that valylthreonine would be a different dipeptide
Polypeptides
- peptides are abbreviated with 3-letter codes (sometimes 1-letter
codes)
- note the huge variety of possible polypeptides, using just the 20
common amino acids as building blocks
20 amino acids
400 dipeptides
8,000 tripeptides
160,000 tetrapeptides
- typical proteins have 100 or more amino acids, so the variety is
immense
- note also the variety of possible functional groups available in
those 20 amino acids
- peptides have evolved with a complete array of properties useful
for life processes
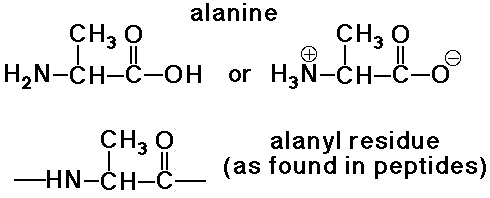
Amino Acid Residues
- an amino acid residue differs from an amino acid by H2O
Disulfide Linkages
- cysteine is readily crosslinked to another cysteine by a S-S bond

- disulfide linkages can hold together parts of a peptide chain that
are not necessarily close in the peptide sequence
Amino Acid Analysis
- primary structure - the order of amino acids in the peptide chain
- complete hydrolysis (extended heating in aqueous HCl) breaks all
amide bonds and releases all the individual amino acids
- individual amino acids can be separated by electrophoresis and/or
chromatography and detected with ninhydrin (purple spot appears on
reaction with any alpha-amino acid)
- complete hydrolysis, separation, and ninhydrin analysis can give
a count of all amino acids and their relative abundance in the peptide
Partial Hydrolysis of Peptides
- occasionally, partial hydrolysis is carried out and smaller peptides
are isolated, which are easier to analyze completely
- from the smaller peptides, sometimes useful connection information
can be determined
- chymotrypsin and trypsin (digestive enzymes) are often used because
they cleave peptides selectively
- chymotrypsin cleaves a peptide only on the carboxyl side of
the aromatic amino acids (Phe, Tyr, Trp)
- trypsin cleaves a peptide only on the carboxyl side of the
strongly basic amino acids (Lys and Arg)
Peptide Sequencing
- complete sequencing of a peptide can be done for up to about 20
amino acids in a row
- Edman degradation: selective cleavage of one amino acid off the
N-terminus, followed by analysis to see what amino acid was removed
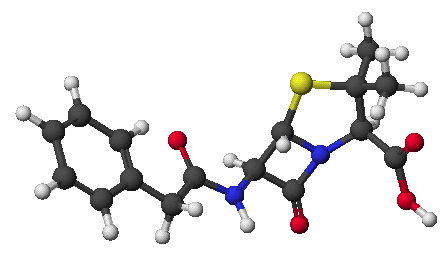
(the selective reagent is phenyl isothiocyanate, and the clipped-off product
is a phenylthiohydantoin)
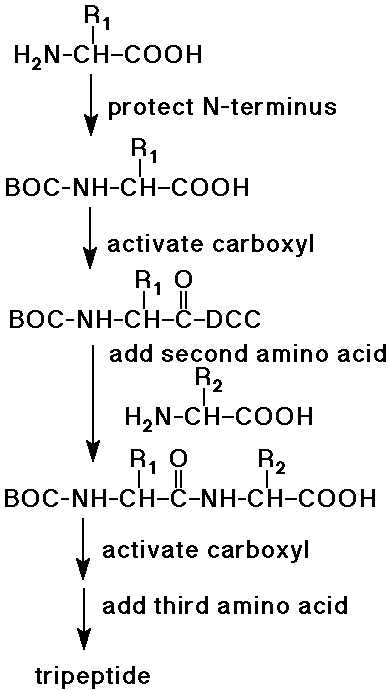
Peptide Synthesis
- complete synthesis of peptides was started by Emil Fischer
- the procedure now can be automated up to about 100 amino acids
- protecting groups are essential to control the coupling reactions
- protect the N-side of AA1
(e.g., as a BOC-amide - easily removed later)
- activate the C-side of AA1
(e.g., with DCC - milder than SOCl2)
- couple active AA1 to AA2
to form a dipeptide (AA2-AA1)
(note that the N-terminus is still protected)
- activate the free carboxyl group of the dipeptide, add AA3
(forms the tripeptide, AA1-AA2-AA3)
- repeat for each amino acid of the polypeptide
- eventually remove the protecting group from the N-terminus
Proteins - Natural Polypeptides
- protein classifications may be based on structure or function or
origin
- proteins are always a polypeptide (sometimes more than one chain)
and may also have other types of molecules associated
Conjugated Proteins are asociated with other types of molecules
- glycoproteins - sugars
- lipoproteins - lipids (nonpolar fats and oils)
- nucleoproteins - nucleic acids
- heme proteins - porphyrin molecule
- metalloproteins - metal ions (often in a heme group)
Protein Shapes
- fibrous proteins - side-by-side polypeptide chains
usually water-insoluble, mechanically strong
useful for structure, muscle
- globular proteins - compact, variable shapes
usually water-soluble, transported around the body
useful for specific functions like catalysis (enzymes)
Protein Functions
- enzymes - catalysts for controlling rates of specific reactions
- hormones - regulators of specific body processes
- transport - control of movement of other molecules or ions
- structure - muscle, skin, etc.
- storage - nutrient storage
- protection - antibodies
Protein Structure - Primary Structure
- amino acid sequence
- determination of the specific order of amino acid residues
Protein Secondary Structure
- regular conformations of the peptide chain
- alpha-helix - a coil held together by H-bonding
- beta-sheet (or pleated sheet) - antiparallel sections of peptide
chains held together by H-bonding
Protein Tertiary Structure
- complete 3-dimensional structure of the protein
- includes disulfide bridges, ionic interactions, H-bonding, other
polar interactions and nonpolar (hydrophobic) interactions
Protein Quaternary Structure
- aggregation of several peptides chains into a larger protein unit
Enzymes - Protein Catalysts
- enzymes affect the rate of a particular reaction
usually very specific as to what reaction or family of reactions will be
catalyzed
- nomenclature: -ase suffix after a description of the reaction they
catalyze
- hydrolase - hydrolysis
- isomerase - isomerization
- transferase - transfer of a group from one molecule to another
- lyase - elimination or addition of a small molecule, like
water
- oxidoreductase - oxidation or reduction (usually with NADH
or similar cofactor)
- ligase - binding of two molecules (usually with ATP cofactor)
- note that enzymes can catalyze a reaction in both directions
like any catalyst, they provide a lower-energy pathway between reactants
and products
actual direction depends on the concentrations of substrates and cofactors
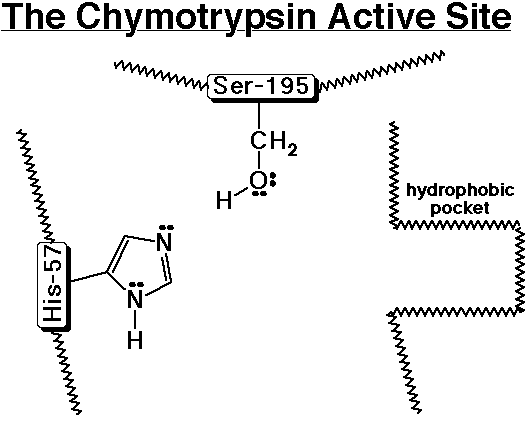
Chymotrypsin - A Specific Example of a Proteolytic Enzyme
- chymotrypsin cleaves an amide bond next to an aromatic amino acid
- the imidazole ring of a nearby histidine specifically adds or removes
a proton to aid in nucleophilic attack or leaving group departure
![]()

![]()
![]()

![]()
![]()

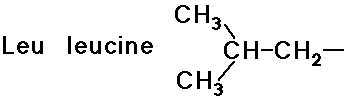
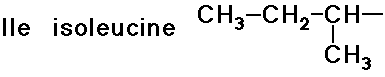


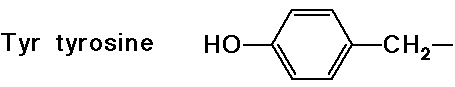

![]()
 (also
note that tyrosine is a phenol)
(also
note that tyrosine is a phenol)![]()
![]()
![]()
![]()
![]()
![]()
![]()