Report on the structure
of the peptide, D_12_N mutant.
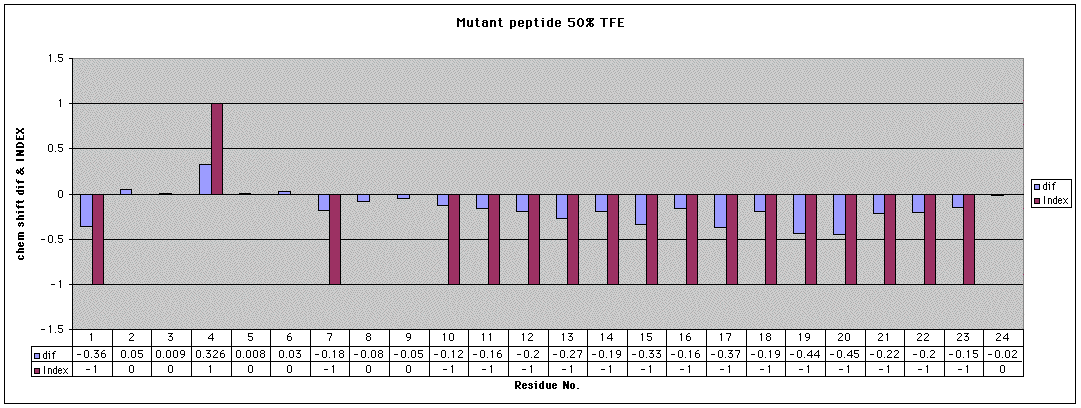
First, here is an analysis of the Ha
chemical shifts (resonance positions vs. known ‘random coil’ positions).
In this analysis, the negative values indicate likely helical regions.
This analysis strongly shows the presence of a helical region from about
residue 10 to residue 21.
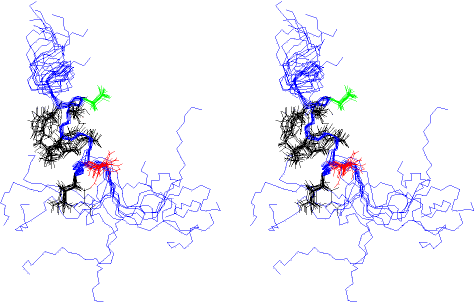
Next is an overlay stereo view of a few (12) overlaid
structures. In this view, Met_1 is at the bottom. I show the side chains
of Asn_12 (red) and Asp_16 (green).
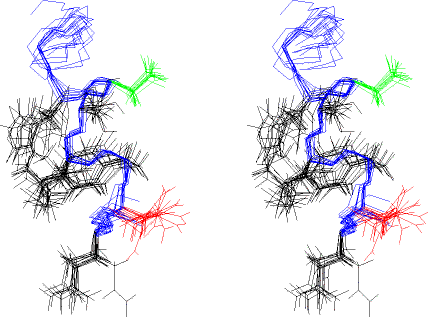
Next, is an expanded region from the above set of
structures (residues 10 — 20), with the side chains from residues 11 -
17 shown. The orientation and colors are as in the previous drawing. This
is also a stereo view.
Next I show a stereo view of one of the structures,
in the same orientation as above. (For my reference, this is structure
‘accept_10.pdb’.)
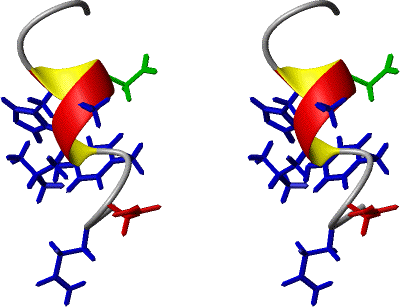
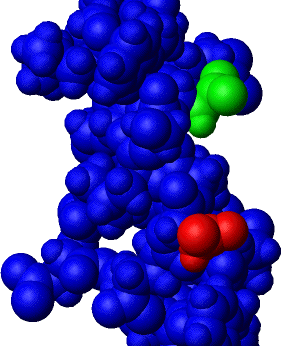
Finally, and not in stereo, here is the same structure
as above, but with van der Waals radii placed on all the atoms. Only residues
12 (red) and 16 (green) are not blue.
So, what does the above mean?
First, the NMR data is of sufficient quality to render
structural data, at least in 50% trifluoroethanol (TFE) co-solvent, the
condition under which the spectra were recorded.
Second, the side chains from residues 12 and 16 pear
to be within a helical region, or at least residue 16 is; residue 12 is
at the end, or near the end. And so the side chains of residues 12 and
16 appear to be situated proximal to each other, on one side of the helix.
As to which direction(s) these side chains ‘face’,
this is probably not defined in such as small, linear peptide as this.
I also have strong doubt as to the structure is so well defined when in
pure water (where there is some solubility problem as well). On the other
hand, the possible binding of an Arg between Asp's 12 & 16 might tie
together (or solidify) this region of the peptide.