Data Structures
Since DNA data contains extremely large volumes of data, efficient data structures are required.
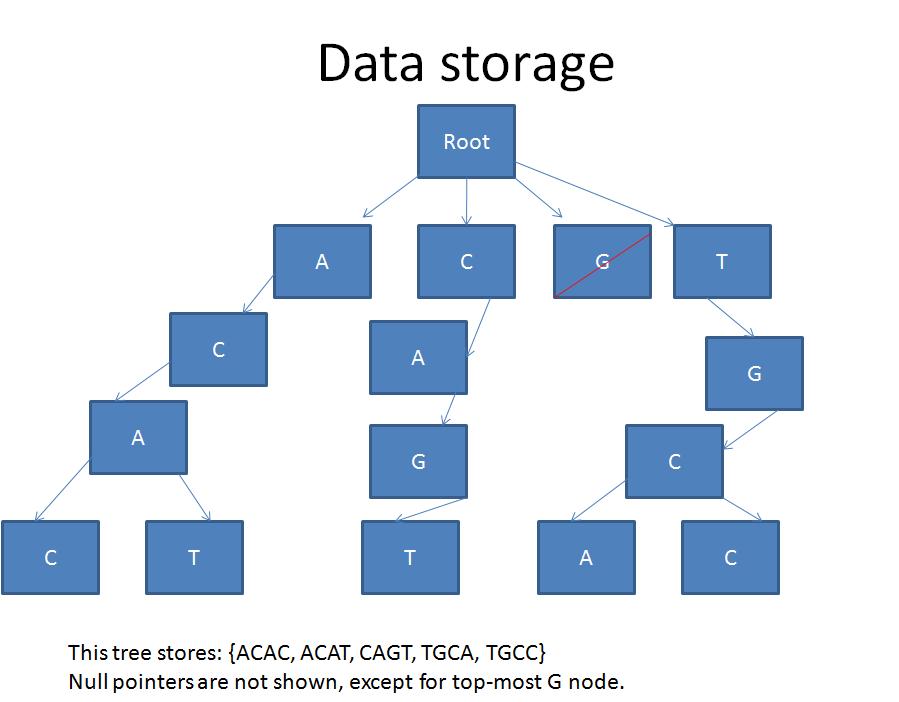
Currently, we are using the following tree structure, however, we are investigating more efficient ways of storing large amounts of data.
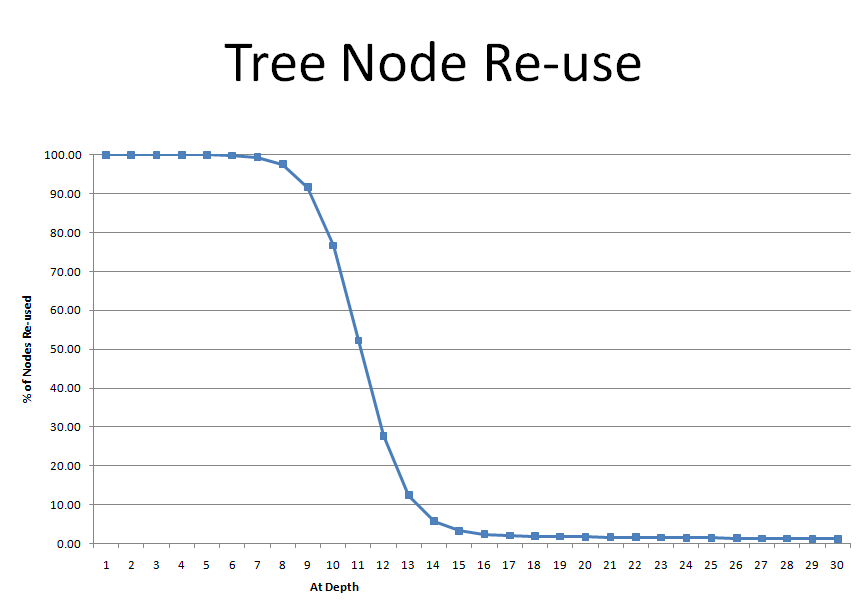
Commonality in the beginning of the sub-strings results in some memory savings due to re-use of tree nodes at the top layers of the tree. Layers further down into the tree have less re-use.
// A node for a linked list of matches found in the DNA file for a given sub-string
struct matchList_str {
int matchposition;
matchList_str *next;
};
// Node in tree (i.e. the rectangle in the previous slide.)
struct node {
struct node *anext;
struct node *cnext;
struct node *gnext;
struct node *tnext;
struct matchList_str *matchList;
};

The Chart below shows that one string has a >90% chance of matching some other string in the genome in the first 9 characters. These "hits" in the tree help reduce the amount of memory required as the already existing nodes can just be followed in the tree w/o needing new nodes to be created.